Abstract:
In recent decades, signal crayfish have rapidly spread across Japan from only three founding populations. To infer the history and explore the success of this signal crayfish invasion, we combined detailed phylogeographical and morphological analyses conducted in both their introduced and native ranges. A combination of high genetic diversity in older populations of the invasive range and rapid adaptation has manifested as larger chela in newer invasions, likely contributing to invasion success in Japan.
The signal crayfish (Pacifastacus leniusculus) is among the world’s most notorious freshwater invaders, which has negatively affected biodiversity and ecosystem services through predation, competition, ecosystem engineering and transmission of diseases (Figure 1). Native to the Pacific Northwest region of North America (northwest United States and southwest Canada), the signal crayfish has been introduced to 27 countries or regions in Europe and Japan for aquaculture. In Japan, signal crayfish were imported five times between 1926 and 1930 from the Columbia River basin in the Pacific Northwest by the former Ministry of Agriculture and Forestry. Earlier studies using ectosymbiont crayfish worms (Branchiobdellida (Annelida)) and polymorphic microsatellite DNA markers revealed that signal crayfish in Japan consist of three founding populations: Hokkaido (Lake Mashu), Shiga (Tankai Reservoir) and Nagano (an irrigation stream in Akashina Town). Nevertheless, the detailed geographic and phylogenetic origins of the invasive signal crayfish in Japan were largely unknown. To infer invasion history of signal crayfish in Japan, both native and introduced ranges need to be studied and contrasted.
The literature suggests that some species may succeed in the invasion process owing to rapid adaptation, evolutionary change or phenotypic plasticity. In crayfish, chela size is highly associated with aggression, dominance and competitive ability, all of which are beneficial when resource availability is limited. Therefore, chela size is considered an important factor that determines their invasion success. A comparison of native and invasive range signal crayfish populations provided an opportunity to also evaluate whether potential invasive traits like chelae size show changes following invasion and secondary spread.
In this study, a research team, consisting of seven universities or institutes from three countries (Japan, United States and Canada), used a large genetic dataset from crayfish sampled in both native and introduced ranges to investigate the invasion history of the signal crayfish in Japan, and made morphological comparisons between these native and introduced populations.
Through mitochondrial DNA analysis (based on16S rRNA gene sequencing), the research team found 15 different haplotypes in the 20 introduced populations of signal crayfish in Japan and 69 haplotypes in the 50 native range populations in North America (Figure 2). The researchers also found that the introduced signal crayfish populations in Japan originate from multiple source populations from the most widely-distributed genetic group in the native range, encompassing British Columbia, Washington, Oregon, Idaho and northern Nevada. As a consequence of genetic admixture in older populations in Japan, genetic diversity of the introduced signal crayfish was as high as genetic diversity of the putative native range populations. However, there was a significant negative relationship between the number of haplotypes and the year of establishment or discovery in the Hokkaido signal crayfish, indicating reduced genetic diversity following secondary spread in this region (Figure 3A).
The research team also found that signal crayfish in Japan have larger chela relative to their putative source populations in the native range (Figure 3B). Furthermore, there was a positive relationship between chela size and the year of establishment or discovery in Hokkaido, suggesting rapid adaptation following secondary spread for these crayfish (Figure 3C).
In sum, the research team established that invasive signal crayfish in Japan have multiple native range source populations, and that high genetic diversity associated with this admixture in older invasive populations in Hokkaido attenuates to lower genetic diversity in younger populations associated with secondary spread or subsequent introductions within the country. Chela size, which is associated with aggressive behaviour and competitive dominance in crayfish, tends to be larger in the invasive range than native range for signal crayfish, and has a tendency to become larger in newer relative to older populations within Japan. A combination of high genetic diversity, especially for older populations in the invasive range, and rapid adaptation to colonisation, manifested as larger chela in recent invasions, likely contributes to invasion success of signal crayfish in Japan.
Future studies should address mechanisms of potential rapid adaptation within this species to advance broad understanding of success of signal crayfish invasions globally and develop management strategies for this and other freshwater invaders.

Figure 1. Photo of signal crayfish (Pacifastacus leniusculus) (© Nisikawa Usio).
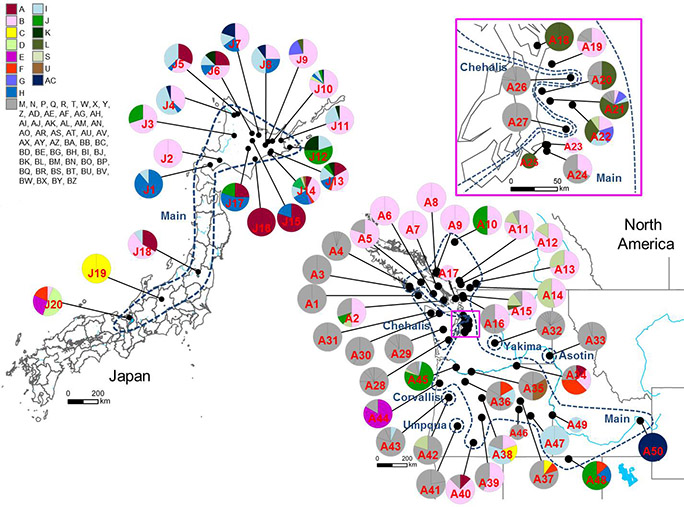
Figure 2: Geographic distribution and haplotypes observed in signal crayfish (Pacifastacus leniusculus) populations in Japan and North America. Smaller circles (A17, A23, A25, A46 and A49) indicate smaller sample sizes (< 3 individuals). The genetic groups of the introduced and native range populations are delineated by blue broken lines. Haplotypes found in Japan are in colour, whereas those found only in North America are in grey.
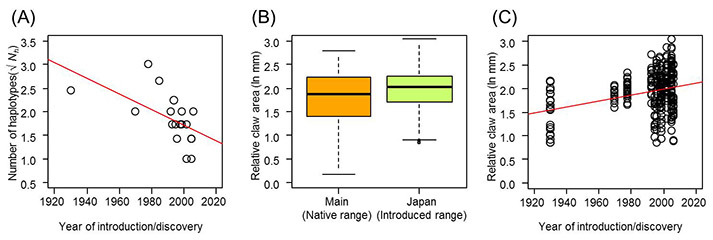
Figure 3: (A) The relationship between the number of mitochondrial DNA haplotypes (Nh) and the year of introduction or discovery for each signal crayfish population of the Hokkaido group (y = −0.0167√x + 35.057, r = −0.581, P = 0.011). (B) Box plots of relative claw area (in natural-log mm) of signal crayfish in the native-range Main group (i.e. putative source populations) and Japan (introduced populations) (linear mixed effects model: t = 2.047, P = 0.049). Thick lines indicate medians and the boxes show the interquartile ranges. Whiskers denote 1.5 times the interquartile ranges. (C) The relationship between relative claw area (in natural-log mm) and the year of introduction or discovery in the Hokkaido group (linear mixed effects model: t = 2.141, P = 0.049).
Article
Title: Phylogeographic insights into the invasion history and secondary spread of the signal crayfish in Japan
Journal: Ecology and Evolution
Authors: Nisikawa USIO1, Noriko AZUMA2, Eric R. LARSON3, Cathryn L. ABBOTT4, Julian D. OLDEN5, Hiromi AKANUMA6, Kenzi TAKAMURA7 & Noriko TAKAMURA7
1Kanazawa University, Japan, 2Hokkaido University, Japan, 3University of Illinois, USA, Fisheries and Oceans Canada, Canada, 5University of Washington, USA, 6Niigata University, Japan, 7National Institute for Environmental Studies, Japan
Doi: 10.1002/ece3.2286
Funder
Grants-in-aid for Scientific Research from JSPS



 PAGE TOP
PAGE TOP