Researchers at Kanazawa University report in Nano Letters how high-speed atomic force microscopy can be used to assess the effectivity of spike-neutralizing antibodies for preventing COVID-19. The use of such antibodies offers a promising alternative to vaccines.
Vaccines against coronavirus disease 2019 (COVID-19) developed and produced during the COVID-19 pandemic offer a significant degree of protection. However, after the second dose of a vaccine, protection only lasts up to 8 months and requires multiple booster doses afterwards. An alternative pathway to prevention and treatment of COVID-19 lies in so-called spike-neutralizing antibodies (SNABs) that neutralize the effect of the spike protein of the SARS-CoV-2 virus, the protein mediating entry of the virus into a host cell. Being able to properly study the interactions between SARS-CoV-2 spike proteins and SNABs is crucial for developing effective treatment — dynamically visualizing these interactions is particularly desirable. Keesiang Lim from Kanazawa University and colleagues have now succeeded in video-imaging protein–SNAB interactions by means of high-speed atomic force microscopy (HS-AFM). They were able to identify the molecular mechanisms at play, and by doing so demonstrated that HS-AFM offers a unique nanoscopic assessment platform for evaluating the intermolecular binding properties of SNABs.
One of the main reasons for the need to study protein–SNAB interactions is to check that they do not lead to a phenomenon called antibody-dependent enhancement (ADE), which refers to the situation when a virus enhances its entry potential by binding to a ‘suboptimal’ antibody. Lim and colleagues used HS-AFM to investigate the ADE risk, as the method is a powerful nanoimaging tool for visualizing molecular structures and the dynamics of biochemical interactions involving viruses.
The scientists first looked at how a SARS-CoV-2 virus spike protein binds to a SNAB. They identified how the structure of the protein–SNAB complex blocks infection, and could conclude that, based on the obtained structural information, ADE risk is negligible. Then, to simulate the dynamic interaction between SNABs and variants of the SARS-CoV-2 virus, Lim and colleagues created small extracellular vesicles (‘biocontainers’ surrounded by a membrane, residing outside cells) capable of producing spike proteins. From these experiments, the researchers concluded viral surface roughness can enhance the infectivity of SARS-CoV-2 variants. They also found that SNAB could neutralize the Delta variant spike protein.
The work of Lim and colleagues shows that HS-AFM provides an excellent nanoscopic assessment tool for studying the binding pattern of SNABs and the dynamic interaction between SNABs and spike protein-mimicking extracellular vesicles. Quoting the researchers: “These results are essential for the screening of appropriate SNABs with a lower risk of ADE to treat COVID-19.”
Related image
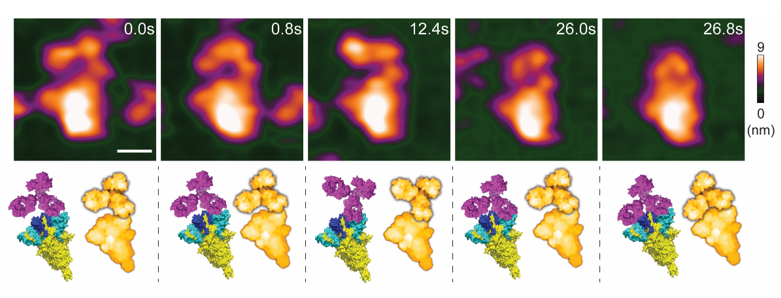
Figure 1.
High-speed atomic force microscopy visualization of a spike-neutralizing antibody reacting with a spike protein.
© 2023 Lim, et al.
Background
High-speed atomic force microscopy
The general principle of atomic force microscopy (AFM) is to make a very small tip scan the surface of a sample. During this horizontal (xy) scan, the tip, which is attached to a small cantilever, follows the sample’s vertical (z) profile, inducing a force on the cantilever that can be measured. The magnitude of the force at the xy position can be related to the z value; the xyz data generated during a scan then result in a height map providing structural information about the investigated sample. In high-speed-AFM (HS-AFM), the working principle is slightly more involved: the cantilever is made to oscillate near its resonance frequency. When the tip is moved around a surface, the variations in the amplitude (or the frequency) of the cantilever’s oscillation — resulting from the tip’s interaction with the sample’s surface — are recorded, as these provide a measure for the local ‘z’ value. AFM does not involve lenses, so its resolution is not restricted by the so-called diffraction limit as in X-ray diffraction, for example.
HS-AFM results in a video, where the time interval between frames depends on the speed with which a single image can be generated (by xy-scanning the sample). Researchers at Kanazawa University have in recent years developed HS-AFM further, so that it can be applied to study biochemical molecules and biomolecular processes in real-time. Keesiang Lim and colleagues have now applied to method to study the process in which a spike-neutralizing antibody binds to the SARS-CoV-2 virus, thereby preventing it from entering a host cell.
Reference
Authors: Keesiang Lim, Goro Nishide, Elma Sakinatus Sajidah, Tomoyoshi Yamano, Yujia Qiu, Takeshi Yoshida, Akiko Kobayashi, Masaharu Hazawa, Toshio Ando, Rikinari Hanayama, and Richard W. Wong.
Title: Nanoscopic Assessment of Anti-SARS-CoV‑2 Spike Neutralizing Antibody Using High-Speed AFM.
Journal: Nano Letters, 2023
DOI: 10.1021/acs.nanolett.2c04270
https://doi.org/10.1021/acs.nanolett.2c04270
Funding
This work was supported by the Grants-in-Aid for Scientific Research (KAKENHI) program of JSPS, Kanazawa University NanoLSI Transdisciplinary Research Promotion Grant, CREST of JST, the Kobayashi International Scholarship Foundation, the Shimadzu Science Foundation, and the Takeda Science Foundation.



 PAGE TOP
PAGE TOP