Abstract:
Current evidence suggests that Southeast Asia was occupied by Hòabìnhian hunter-gatherers until ~4000 years ago, but the human occupation history of Southeast Asia thereafter with farming economies developed and expanded remains heavily debated. By sequencing 26 ancient human genomes (25 from Southeast Asia, 1 Japanese Jōmon), we show that Southeast Asian history is more complex than previously thought; both Hòabìnhian hunter-gatherers and East Asian farmers with further migrations contributed to current Southeast Asian diversity. Our results help resolve one of the long-standing controversies in Southeast Asian prehistory.
[Background]
Uncovering the expansion processes of human habitats in the past is of great importance for understanding the origins and establishment of present-day populations and the acquisition of genetic characteristics of individuals as well as for investigating mechanisms of resistance against diseases and pathogens. Previous genetic/genomic studies aimed to uncover the expansion processes using present-day human genomes of different individuals and locations. However, it is not always possible to elucidate the expansion processes based on the genomic similarity of present-day populations due to the possibility of migrations of populations between regions in various periods. It is therefore impossible to uncover the precise expansion processes of populations in the past without knowledge of the genomic information existing in a designated region and period. Thus, expansion processes hypothesized so far were nothing but speculations based on assumptions about present-day genomes.
Recent developments of DNA analysis technology have made it possible to obtain whole genome information from ultratrace amounts of DNA; we are now in an era where whole genome information can be obtained directly from ancient human skeletons discovered at archaeological sites. There remain, however, technical problems for obtaining whole genome information of ancient human skeletons. In particular, there are two main problems: first, genomic analyses*1) of poorly-preserved ancient remains in hot and humid regions of the world have up until now failed (Figure 1). Secondly, there is the risk of contamination of present-day human DNA in the DNA samples of ultratrace amounts from prehistoric remains. To evaluate objectively the possibility of such contamination, several different research groups must cross-check*2) one another in order to achieve exact genome sequencing; in other words, establishment of a collaborative research system is a prerequisite for attaining the highest level of scientific authenticity.
In order to cope with these problems, the present international research team, led by researchers from the University of Copenhagen with the participation of three researchers from Kanazawa University has established technologies to efficiently extract human DNA from skeletons discovered at prehistoric remains even under very poor conditions for DNA preservation. At the same time, an international system of research collaboration has been established for objectively evaluating the effects of contamination by present-day human DNA. Thanks to these efforts, the team has uncovered the expansion processes of human habitats and genetic interactions in hot and wet Southeast Asia, which was not possible previously with conventional technologies and research systems (Figure 2).
Worthy of special mention, the present study has been successful in determining the “whole genome” sequence of an individual with typical Jōmon culture, while previous studies were only able to show a very limited “partial genome” sequence of two Jōmon individuals. Thus, the present study is the first successful example to show the possibility of whole genome sequencing of prehistoric individuals in regions like Japan where preservation conditions are quite poor, possibly leading to further major progress in prehistoric genome studies.
[Results]
In the present study, the international research team succeeded in extracting and sequencing DNA from 25 ancient individuals’ skeletons from Southeast Asian remains, where the condition of DNA preservation is very poor, and from one Japanese Jōmon female skeleton. Upon comparison of the genomic data of ancient human skeletons with those of present-day human skeletons, it has become clear that those prehistoric populations in Southeast Asia can be classified into six groups (Figure 3).
Group 1 contains Hòabìnhians from Pha Faen, Laos, hunter-gatherers (~8000 years ago), and prehistoric populations discovered from Gua Cha, Malaysia (~4000 years ago), being genetically close to present-day Önge and Jarawa from the Andaman Islands and Jehai from the Peninsular Malaysia. To our surprise, group 1 has higher genetic affinities with Ikawazu*3) Jōmon individual (Tahara, Aichi), a female adult*4), than other present-day Southeast Asians. In addition, the Ikawazu Jōmon genome*5) is best modelled contributing genetically present-day Japanese.
On the other hand, Groups 2-6 consist of ancient skeletons from the Neolithic Age, when farming started, until ~500 years ago. It is now found that they are genetically much different from Hòabìnhians, each group having histories of migration and genetic interaction, i.e., inter-population mixture. Group 2 is found to be genetically close to the present-day Austroasiatic language-speaking groups such as Mlabri, but to have few genetic components common with the present-day East Asian populations. Group 3 is found to be genetically close to Kradai, Thailand, in the present-day Southeast Asian populations and to the Austronesian language-speaking groups. Group 4 is found to be genetically close to the present-day populations in South China. Group 5 is genetically close to the present-day populations in the western part of Indonesia. Group 6 is most closely related to present day Austronesian populations, with one individual showing slightly elevated Denisovan ancestry, an archaic hominin which is classified as a sister group of Neanderthals.
As above, Neolithic Southeast Asians are found to have been partially genetically influenced by ethnic groups in South China and to have had a genetic connection with populations in Taiwan; Neolithic Southeast Asians are found not to have been indigenous hunter-gatherers passively accepting farming but to have accepted farming gradually in the process of migrations of populations between the continent and islands. Conventional archaeology proposed the two-layer hypothesis that, in those periods, a large population with farming culture with rice and millet migrated into Southeast Asia and that they replaced the indigenous population. Additionally, the present study indicates that the genetic influence from South China with rice farming was only partial and that the migrating population did not replace the indigenous population completely. The present analysis shows that there were at least four big migration waves; migrations of Southeast Asians should be investigated with a new “complex model” framework.
The present study successfully elucidates for the first time the expansion/migration of prehistoric populations by genome analysis of skeletons discovered in Southeast Asia; conventionally, it was thought that such population expansion/migration could only be investigated using archaeological artifacts. An important outcome of the present study is that the same or analogous analyses could be applied to various regions to evaluate the history of population expansion/migration in much more detail and in a more scientific manner.
[Future prospects]
The genomic data obtained from ancient skeletons in Southeast Asia and from a Ikawazu Jōmon individual provides an important basis for investigations on the origins of populations in wider East Asia. The whole genome information of a Jōmon individual will be useful for direct comparison of genomic similarity with ancient East Asians of the corresponding period to Jōmon in present-day Korea, China, Russia and others in the vicinity of the Japanese archipelago. More comparative studies are in progress on populations in wider areas. Note that the whole genome sequence obtained in this study for a Jōmon individual corresponds to the Draft Genome Sequence in the Human Genome Project for the present-day humans. We aim at Complete Genome Sequence with higher accuracy.
This study is an interdisciplinary undertaking combining anthropology and archaeology in a close collaboration, allowing us to establish ourselves at the starting point for research on the origin of Jōmon and its diversity. By more genome analyses of more Jōmon skeletons from different Jōmon sites, genetic diversity of Jōmon populations will be explored over the Japanese archipelago. It is expected through such studies that various interactions among Jōmon groups should be revealed together with migrations of archaeological artifacts such as potteries and stone tools as well as migrations of populations. Based on the outcome of the present study, novel anthropological and archaeological approaches would be further developed.
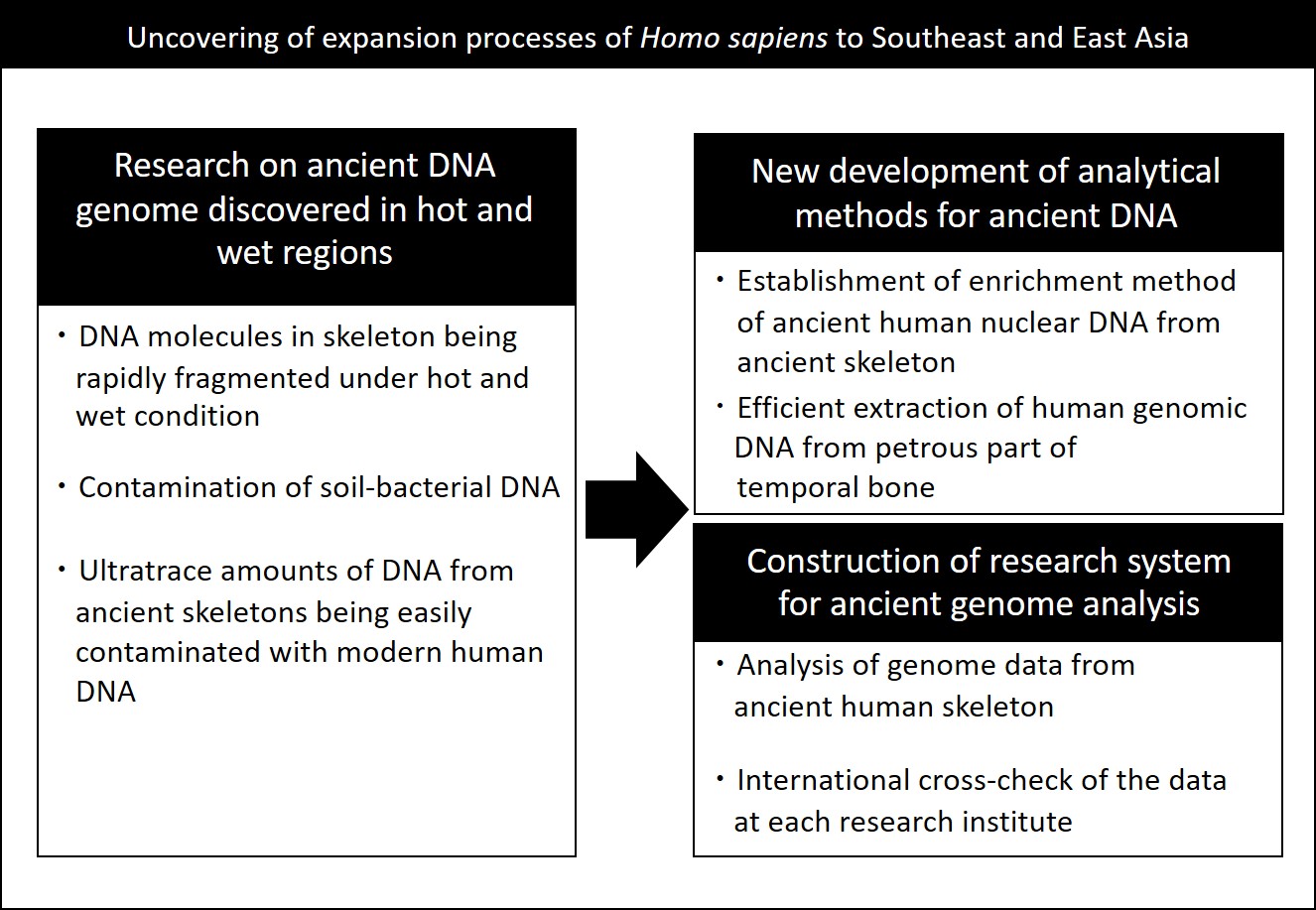
Figure 1. Outline of research background

Figure 2. Experiments concerning genome analysis of ancient skeleton.
(In the clean room specialized for ancient DNA analyses, Kitazato University School of Medicine.)
Top left: DNA extraction in a clean bench under the condition with very little modern human DNA.
Top right: Disposable dustproof lab coats and special rubber gloves only are allowed to use.
Bottom: Sampling from temporal bone of a Jōmon human individual.
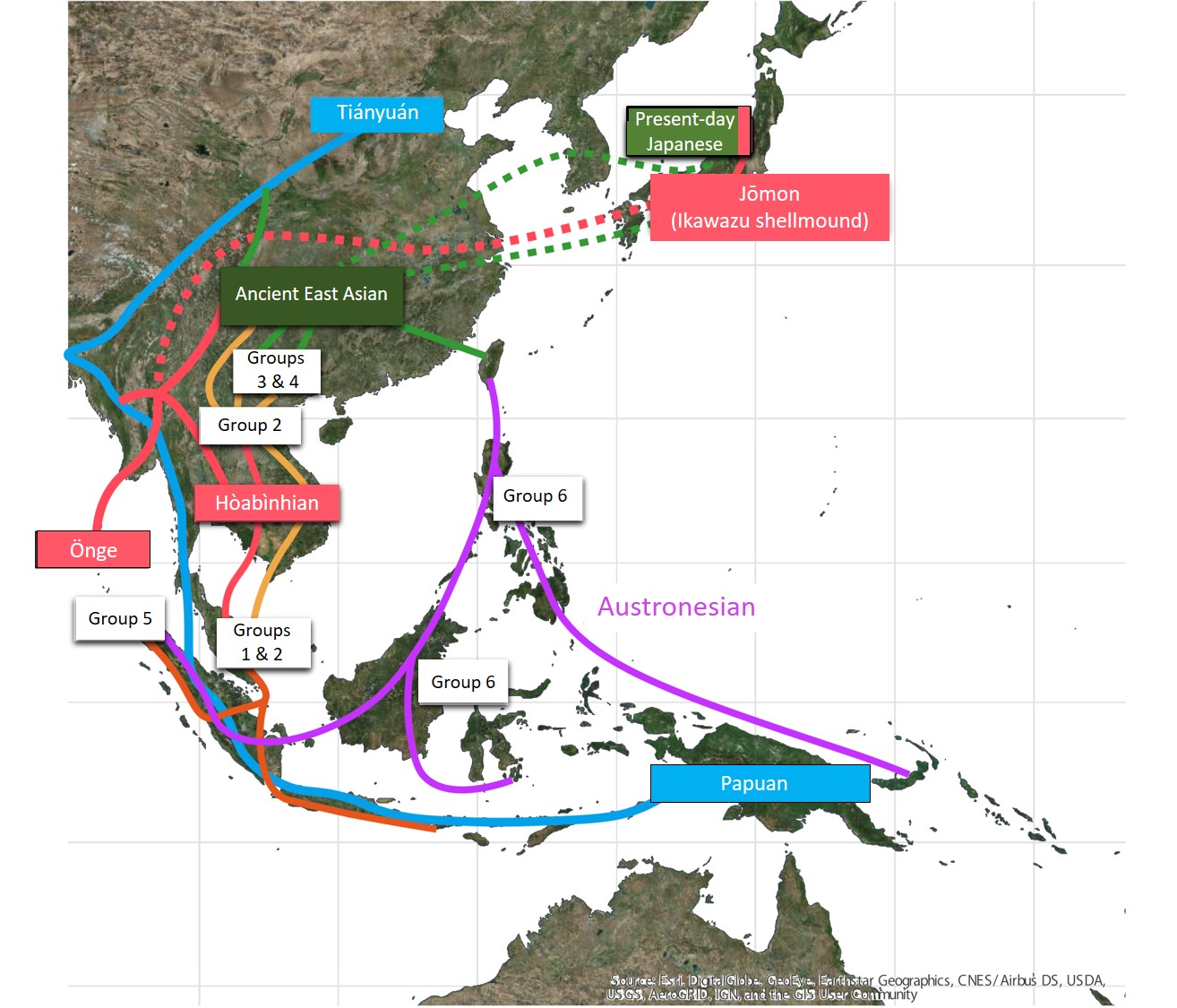
Figure 3. Model for migration routes into Southeast Asia uncovered by genomic data of prehistoric skeletons.
[Glossary]
*1) Genomic Analysis
Analysis of whole genome of a species.
*2) Cross-check analysis
Cross-check analysis is an analytical method that evaluates whether the same result will be produced irrespective of different research institutions, different analytical methods, and so on. It is an important scientific index in research on ancient DNA.
*3) Ikawazu kaizuka (shellmound)
A kaizuka (shellmound) site at Tahara city, Aichi prefecture, dating back to late and final Jōmon period. One of the best known archaeological site of Jōmon period, where more than 200 individual skeletons have been discovered from Meiji era till today. A number of renowned anthropologists like Profs. Yoshikiyo KOGANEI and Hisashi SUZUKI performed morphological research on prehistoric skeletons from this site. There are also other kaizuka sites in Tahara city, such as Yoshigo kaizuka and Hobi kaizuka, representative Jōmon sites. Those sites have been well studied and many skeletons have been excavated.
*4) A female skeleton dating back to late Jōmon period, ~2500 years ago.
A Jōmon skeleton discovered from Ikawazu kaizuka site in 2010. Recent studies indicate the beginning of Yayoi period to be ~3000 years ago, but the arrival of Yayoi culture differed depending on regions. The female adult skeleton from Ikawazu kaizuka site is accompanied with a pottery that is validated to date back to the period of Gokanmori type pottery, indicating that the period was still Jōmon at those sites in Atsumi peninsula, Aichi prefecture. In addition, the female skeleton analyzed here shows typical Jōmon morphology.
*5) Whole genome sequence
Whole genome sequence is the total DNA sequence of a species covering not only DNA sequences for genes but also DNA sequences for non-gene regions.
Article
The prehistoric peopling of Southeast Asia
Journal: Science
Authors: Hugh McColl, Fernando Racimo, Lasse Vinner, Fabrice Demeter, Takashi Gakuhari, J. Víctor Moreno-Mayar, George van Driem, Uffe Gram Wilken, Andaine Seguin-Orlando, Constanza de la Fuente Castro, Sally Wasef, Rasmi Shoocongdej, Viengkeo Souksavatdy, Thongsa Sayavongkhamdy, Mohd Mokhtar Saidin, Morten E. Allentoft, Takehiro Sato, Anna-Sapfo Malaspinas, Farhang A. Aghakhanian, Thorfinn Korneliussen, Ana Prohaska, Ashot Margaryan, Peter de Barros Damgaard, Supannee Kaewsutthi, Patcharee Lertrit, Thi Mai Huong Nguyen, Hsiao-chun Hung, Thi Minh Tran, Huu Nghia Truong, Giang Hai Nguyen, Shaiful Shahidan, Ketut Wiradnyana, Hiromi Matsumae, Nobuo Shigehar, Minoru Yoneda, Hajime Ishida, Tadayuki Masuyama, Yasuhiro Yamada, Atsushi Tajima, Hiroki Shibata, Atsushi Toyoda, Tsunehiko Hanihara, Shigeki Nakagome, Thibaut Deviese, Anne-Marie Bacon, Philippe Duringer, Jean-Luc Ponche, Laura Shackelford, Elise Patole-Edoumba, Anh Tuan Nguyen, Bérénice Bellina-Pryce, Jean-Christophe Galipaud, Rebecca Kinaston, Hallie Buckley, Christophe Pottier, Simon Rasmussen, Tom Higham, Robert A. Foley, Marta Mirazón Lahr, Ludovic Orlando, Martin Sikora, Maude E. Phipps, Hiroki Oota, Charles Higham, David M. Lambert, Eske Willerslev
Doi: 10.1126/science.aat3628
Funders
This work was supported by the Lundbeck Foundation, the Danish National Research Foundation, and the KU2016 program. H.Mc. is supported by the University of Adelaide’s George Murray Scholarship. R.S. thanks the Thailand Research Fund (TRF) for support (grants RTA6080001 and RDG55H0006). The excavation of the Jōmon individual was supported by a Grant-in- Aid for Scientific Research (B) (25284157) to Y.Y. The Jōmon genome project was organized by H.I.; as well as T.H. and H.O., who were supported by MEXT KAKENHI grants 16H06408 and 17H05132; and a Grants-in-Aid for Challenging Exploratory Research (23657167) and for Scientific Research (B) (17H03738). The Jōmon genome sequencing was supported by JSPS KAKENHI grant 16H06279 to A.T. and partly supported by the CHOZEN project in Kanazawa University and the Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University. Computations for the Jōmon genome were partially performed on the NIG supercomputer at ROIS National Institute of Genetics. M.M.L. is supported by the ERC award 295907. D.M.L. was supported by ARC grants LP120200144, LP150100583, and DP170101313. A.P. is supported by Leverhulme Project Research grant RPG-2016-235. M.E.P. acknowledges the Cardio-Metabolic research cluster at Jeffrey Cheah School of Medicine & Health Sciences, TMB research platform, Monash University Malaysia, and MOSTI Malaysia for research grant 100-RM1/BIOTEK 16/6/2B. A.S.M. was financed by the European Research Council (starting grant) and the Swiss National Science Foundation.



 PAGE TOP
PAGE TOP