Abstract:
In a recent study published in BBA – General Subjects, Kanazawa university researchers have used high-speed microscopy to investigate native structure and conformational dynamics of hemagglutinin in influenza A.
The influenza A viruses, which have instigated deadly pandemics in the past, still remain a major global public health problem until today. Molecules known as virulence factors are produced by bacteria, viruses, and fungi to help them to infect host cells. One of virulence factors found in the influenza A viruses is hemagglutinin (HA). Researchers at Kanazawa University have recently studied the structure of HA of avian influenza virus, H5N1, using high-speed atomic force microscopy (HS-AFM) and the findings are essential for developing therapeutic approaches against influenza A viruses in future.
HA is initially synthesized by host cells in its precursor form that known as HA0. Conversion of HA0 to HA is depending on the pathogenicity of influenza A viruses: extracellular conversion for low pathogenic influenza A viruses and intracellular conversion for highly pathogenic influenza A viruses. Therefore, understanding the structure and properties of HA0 is paramount to deciphering HA. Richard Wong and his research team thus sought out to scrutinize HA0 under the microscope. Recombinant HA0 protein of H5N1 was visually analyzed by HS-AFM system developed by Kanazawa University.
Both HA0 and HA exist in homotrimeric forms and conversion of HA0 to HA does not significantly modify the homotrimeric structure. Therefore, it is sensible to use HA as a template to generate HA0 HS-AFM simulation images. Acidic endosomal environment is the critical factor for HA to induce fusion between viral membrane and endosomal membrane in order to release viral materials into host cells. To elucidate the acidic effect on HA0, it was first exposed to an acidic environment. The trimer of HA0 turned out to be very sensitive to the acidic solution and expanded considerably. When conformational changes of hemagglutinin were measured in real-time using HS-AFM, the team found that its area was larger, and its height shorter. Acidic environment essentially made the molecule flatter and more circular, as compared to its original counterpart. This change in conformation was, however, reversible as the structure reverted back to its original form upon neutralization.
“Our pilot work establishes HS-AFM as an inimitable tool to directly study viral protein dynamics, which are difficult to capture with low signal-to-noise techniques relying on ensemble averaging, such as cyro-EM and X-ray crystallography,” says lead author of the study Dr Kee Siang Lim. “With high scanning speed and a minimally invasive cantilever, we predict that HS-AFM is feasible to reveal the flow of irreversible conformational changes of HA2 induced by low pH, which is mimicking the true biological events that occur when HA enters a host endosome, in future study.”
This study paved the way for investigating biological events within viruses in real-time. The authors state the importance of HS-AFM for this research: “Our work establishes HS-AFM as an inimitable tool to directly study viral protein dynamics, which are difficult to capture with low signal-to-noise techniques relying on ensemble averaging, such as cyro-EM and X-ray crystallography,” explains Dr Richard Wong, senior author of the study.
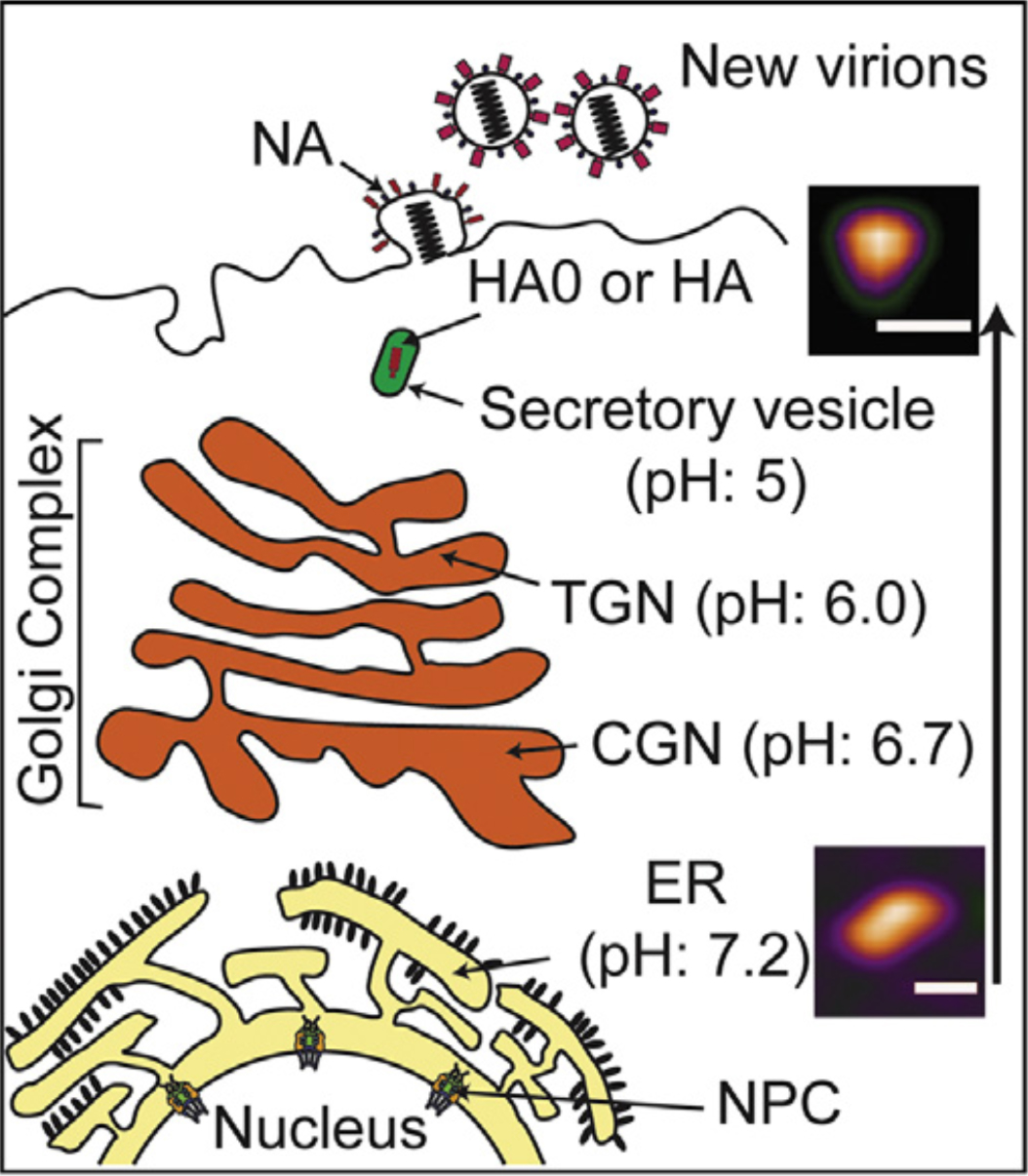
Figure 1.
Low pH environment induces conformational change of HA0. Conformational change of HA0 trimer in neutral and acidic condition mimics to the pH gradient from ER to secretory vesicles during viral assembly in cell membrane (ER: endoplasmic reticulum; CGN: cis-Golgi network; TGN: trans-Golgi network; NPC: nuclear pore complex).
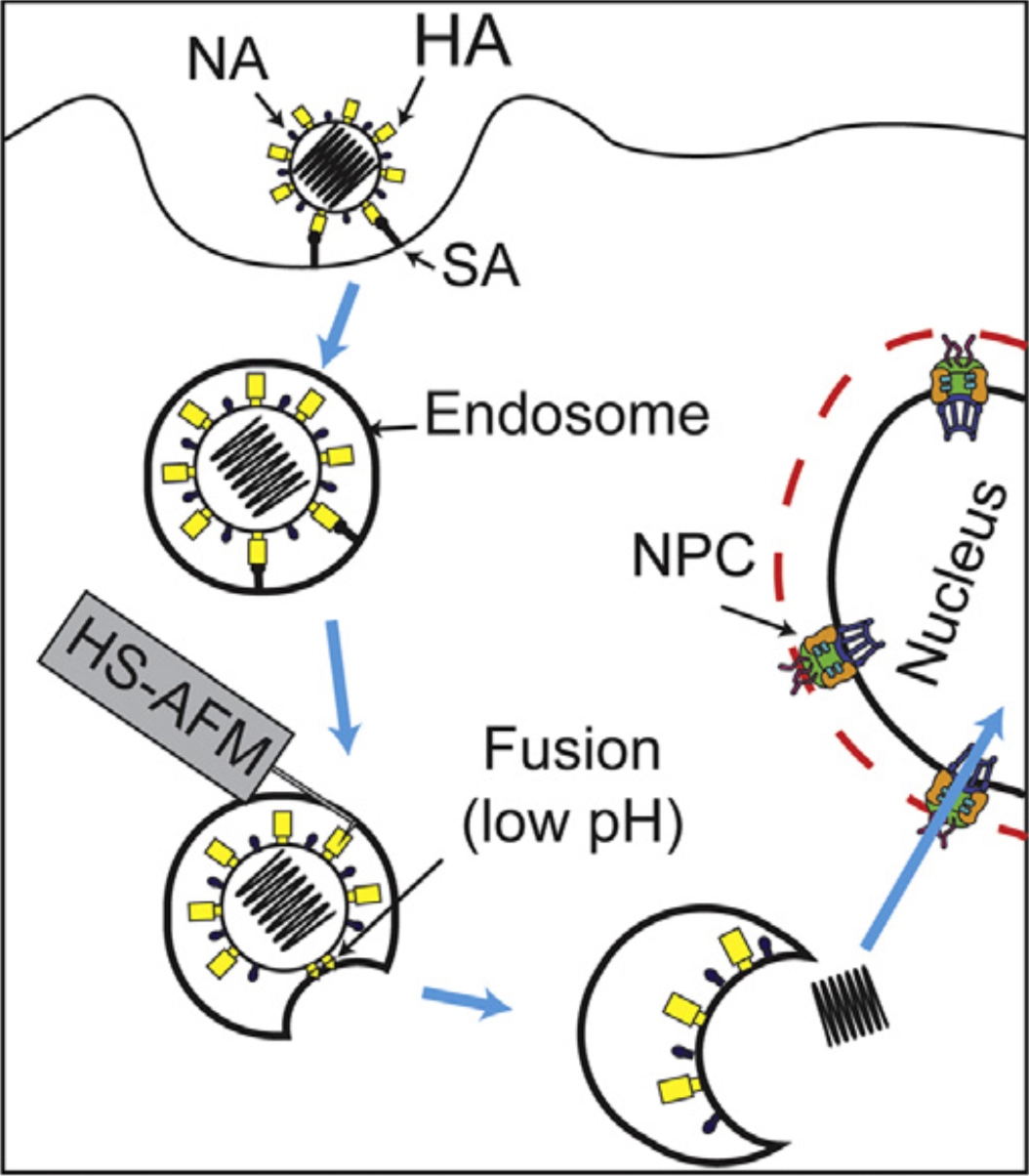
Figure 2.
Strategy for direct visualization of complex and highly dynamic HA structure during viral entry in near future (HA: hemagglutinin, NA: neuraminidase, NPC: nuclear pore complex).
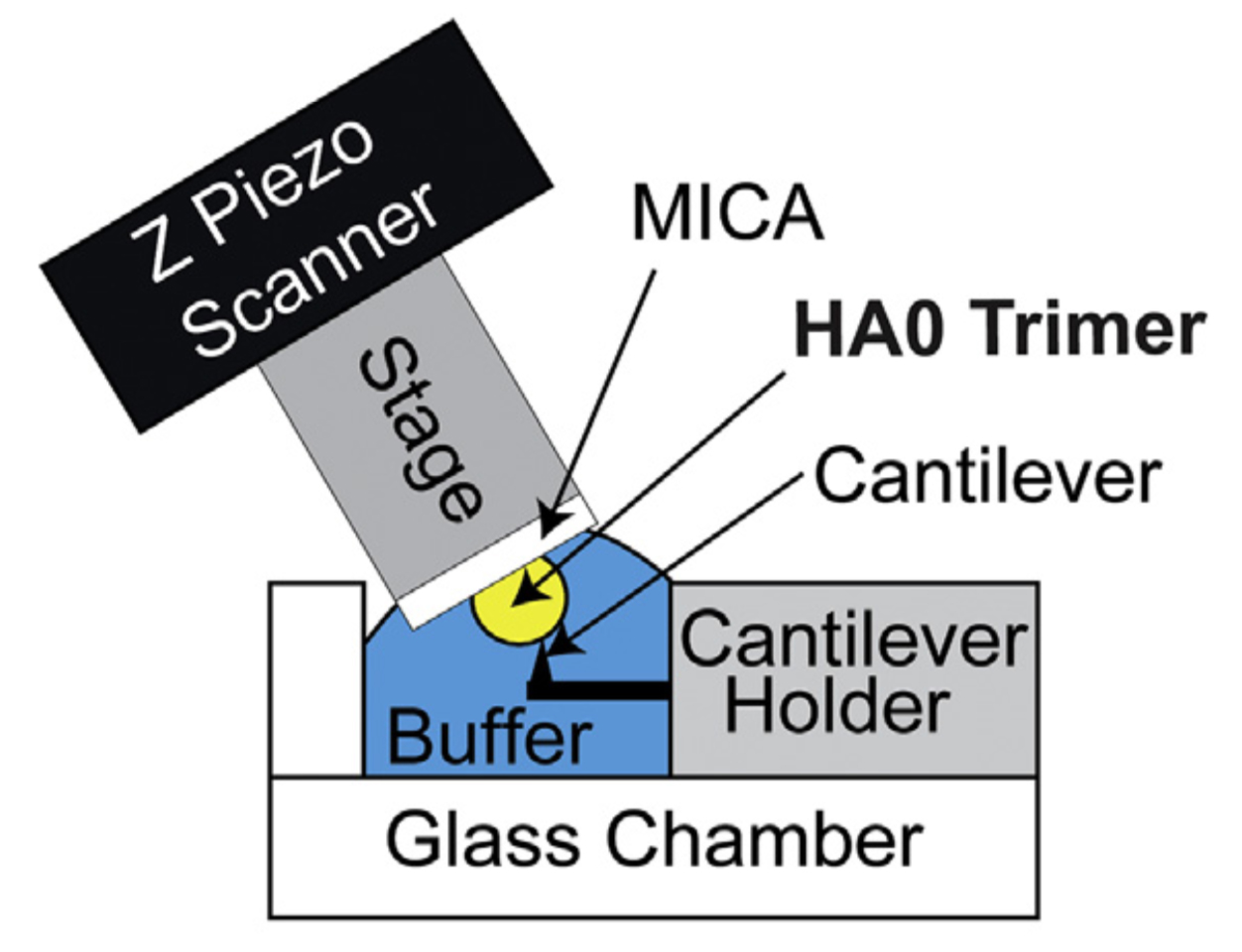
Figure 3.
HS-AFM setup for direct visualization of HA0 trimer. Schematic diagram of the HS-AFM setup for scanning the HA0 trimer.
Article
Direct visualization of avian influenza H5N1 hemagglutinin precursor and its conformational change by high-speed atomic force microscopy
Journal: Biochimica et Biophysica Acta (BBA) – General Subjects
Authors: Kee Siang Lim, Mahmoud Shaaban Mohamed, Hanbo Wang, Hartono, Masaharu Hazawa, Akiko Kobayashi, Dominic Chih-Cheng Voon, Noriyuki Kodera, Toshio Ando, Richard W. Wong
DOI: 10.1016/j.bbagen.2019.02.015
Funder
This work was supported by a WPI NanoLSI start-up fund (to K.L.); MEXT/JSPS KAKENHI grant numbers 17H05874 and 17K08655 (to R.W.) from MEXT Japan, and by grants from the Kobayashi International Scholarship Foundation (to R.W), and the Shimadzu Science Foundation (to R.W.).



 PAGE TOP
PAGE TOP