Abstract:
The function of protein machines in biological cells is so complex that even supercomputers cannot predict their cycles at atomic detail. But, as demonstrated in this review article, many aspects of their operation at mesoscales can be already revealed by exploring simple mechanical models, amenable for simulations on common computers. The authors also show how artificial protein-like structures with machine properties can be designed.
The living cell can be viewed as a factory where protein machines are in charge of various processes, such as transport of material inside the cell or operations with other macromolecules like DNA. Their operation is typically fueled by ATP molecules, the major energy carrier in biological cells. The chemical energy gained through ATP hydrolysis is used by a protein machine to cyclically change its shape and thus to perform a particular function. Hence, resolving functional conformational changes in proteins is a major challenge, with fundamental importance for understanding and control of biological single-molecule motors and machines.
The complexity of interactions between atoms in a protein machine is so high that even world’s best supercomputers cannot reproduce just one of their operation cycles. In this review article, it is however demonstrated that essential aspects of the operation of such natural nano-devices can be already revealed by exploring very simple mechanical models of proteins, i.e. by treating such macromolecules as elastic networks obtained by connecting particles by a set of elastic springs.
The authors, professor Alexander Mikhailov and assistant professor Holger Flechsig from the Nano Life Science Institute in the Kanazawa University in Japan, argue that elastic networks corresponding to protein machines with functional dynamics have special properties, emerged in the process of biological evolution. Despite an apparent complexity, internal motions in such systems proceed in an ordered manner, as if guided along hidden rail tracks. Thus, a molecular machine behaves similar to macroscopic mechanical devices with highly coordinated movements of their parts. This ensures that the cellular factory can function robustly despite strong fluctuations present at nanoscales.
Using this approach, simulations covering entire operation cycles could be performed and the first molecular movies of protein machines were thus obtained. As an example, Fig.1 from the original 2010 publication by the authors shows how the helicase motor protein of the hepatitis C virus – a principal part of its replication machinery and an important pharmacologic target for antiviral drugs – actively moves along the DNA and mechanically unzips it.
Artificial protein-like structures with machine properties could be moreover designed by running a computer evolution of elastic networks. In Fig. 2 two examples are shown, a designed model machine operating within a biological membrane, and a machine which exhibits allosteric communication.
This review article presents a new perspective in understanding of the complex machinery of biological cells. It also paves the way to novel approaches in the design of artificial nano-machines – the task of high potential for future biotechnological applications.
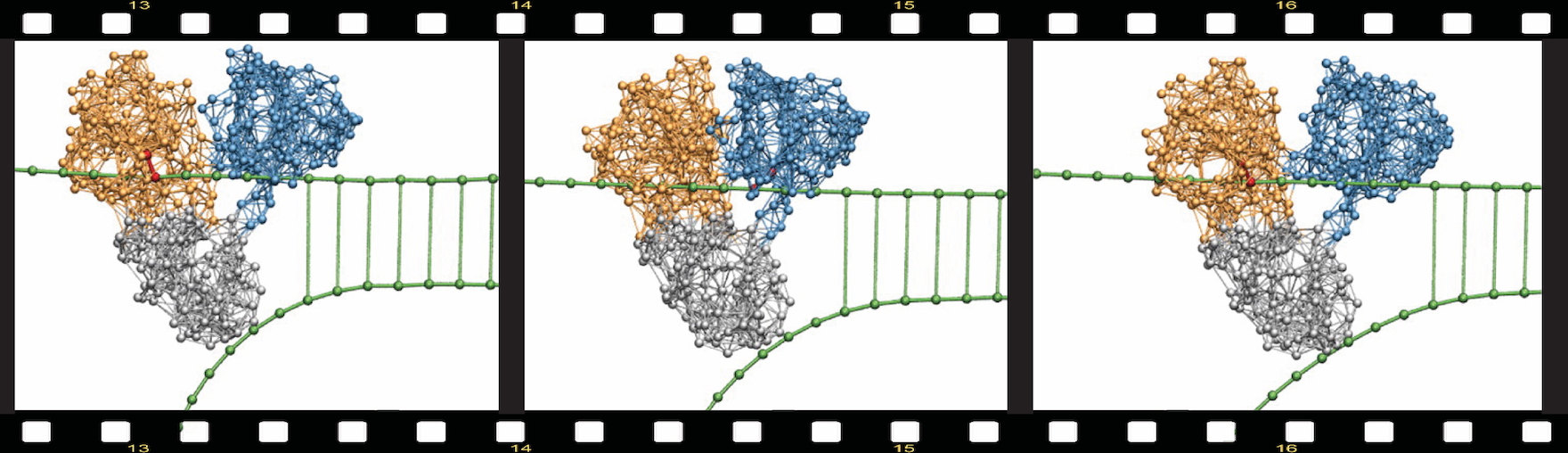
Figure 1. Molecular movie.
Snapshots from a simulation revealing the mechanism by which hepatitis C virus helicase breaks double DNA strands. The two motor domains walk along the upper strand employing the inchworm translocation mechanism. The third domains acts like a wedge mechanically separating the duplex (from Fig. 14).
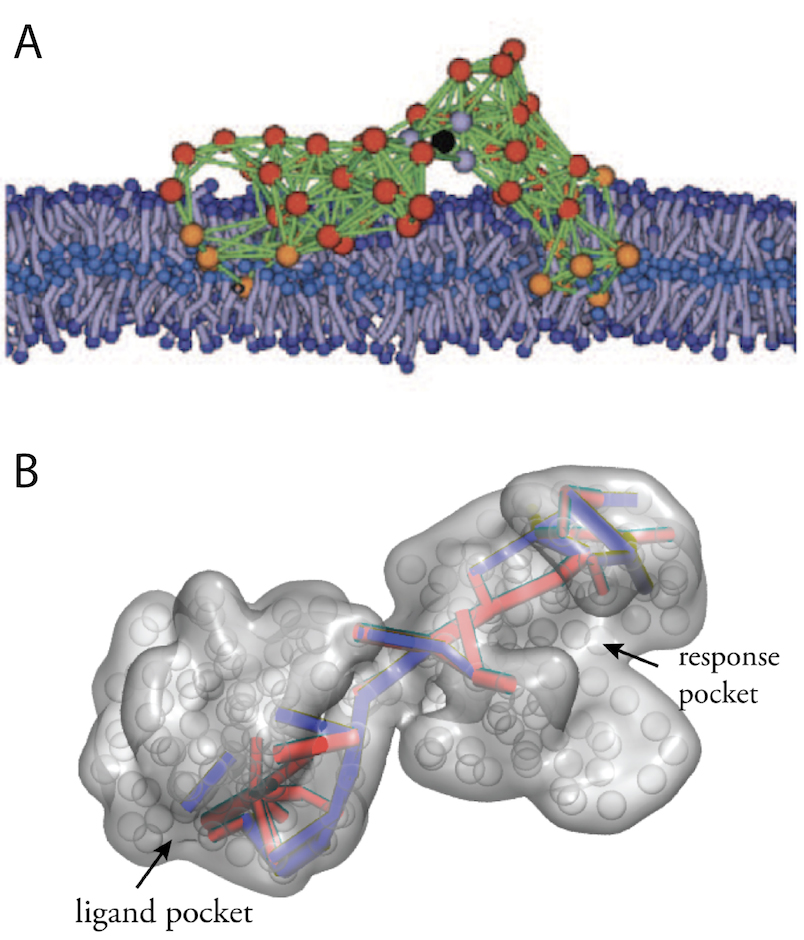
Figure 2. Designed machines.
Artificial protein-like structures obtained by in-silico evolution. (A) A model protein machine in a biological membrane (from Fig. 11). (B) Communication pathways in a designed allosteric structure (from Ref. 34).
Article
Simple mechanics of protein machines.
Journal: Journal of the Royal Society Interface
Authors: Holger Flechsig and Alexander S. Mikhailov
Funder
World Premier International Research Center Initiative (WPI), MEXT, Japan; JSPS KAKENHI grants nos. JP19K03765 and JP16K05518.



 PAGE TOP
PAGE TOP